Question answering using BioBERT
Querying and locating specific information within documents from structured and unstructured data has become very important with the myriad of our daily tasks. Within the healthcare and life sciences industry, there is a lot of rapidly changing textual information sources such as clinical trials, research, published journals, etc, which makes it difficult for professionals to keep track of the growing amounts of information. To find pertinent information, users need to search many documents, spending time reading each one before they find the answer. An automatic Question and Answering (QA) system allows users to ask simple questions in natural language and receive an answer to their question, quickly and succinctly. A QA system will free up users from the tedious task of searching for information in a multitude of documents, freeing up time to focus on the things that matter. Let us look at how to develop an automatic QA system.
Before we start it is important to discuss different types of questions and what kind of answer is expected by the user for each of these types of questions. Generally, these are the types commonly used:
- Factoid questions: Factoid questions are pinpoint questions with one word or span of words as the answer. The answers are typically brief and concise facts. For example: “Who is the president of the USA?”. We will focus this article on the QA system that can answer factoid questions.
- Non-factoid questions: Non-factoid questions are questions that require a rich and more in-depth explanation. For example: “How do jellyfish function without a brain or a nervous system?”
To answer the user’s factoid questions the QA system should be able to recognize the intent behind the questions, retrieve relevant information from the data, comprehend the retrieved documents, and synthesis the answer.
There are two main components to the question answering systems:
- Document Retriever
- Document Reader
Let us look at how these components interact. Figure 1 shows the iteration between various components in the question answering systems.

Dataset used
We will attempt to find answers to questions regarding healthcare using the Pubmed Open Research Dataset. The data was cleaned and pre-processed to remove documents in languages other than English, punctuation and special characters were removed, and the documents were both tokenized and stemmed before feeding into the document retriever.
Document Retriever
The document retriever uses a similarity measure to identify the top ten documents from the corpus based on the similarity score of each document with the question being answered.
The efficiency of this system is based on its ability to retrieve the documents that have a candidate answer to the question quickly. The following models were tried as document retrievers:
- Sparse representations based on BM25 Index search [1]
- Dense representations based on doc2vec model [2]
These models were compared based on the document retrieval speed and efficiency. We experimentally found out that the doc2vec model performs better in retrieving the relevant documents.
Document Reader
The document reader is a natural language understanding module which reads the retrieved documents and understands the content to identify the correct answers. We are using “BioBERT: a pre-trained biomedical language representation model for biomedical text mining” [3], which is a domain-specific language representation model pre-trained on large-scale biomedical corpora for document comprehension. BioBERT is pre-trained on Wikipedia, BooksCorpus, PubMed, and PMC dataset. We fine-tuned this model on the Stanford Question Answering Dataset 2.0 (SQuAD) [4] to train it on a question-answering task. The SQuAD 2.0 dataset consists of passages of text taken from Wikipedia articles. It is a large crowd sourced collection of questions with the answer for the questions present in the reference text. We trained the document reader to find the span of text that answers the question. Our model produced an average F1 score [5] of 0.914 and the EM [5] of 88.83% on the test data. The model is not expected to combine multiple pieces of text from different reference passages. This module has two main functions: the input module and the start and end token classifier.
BioBERT Input Format
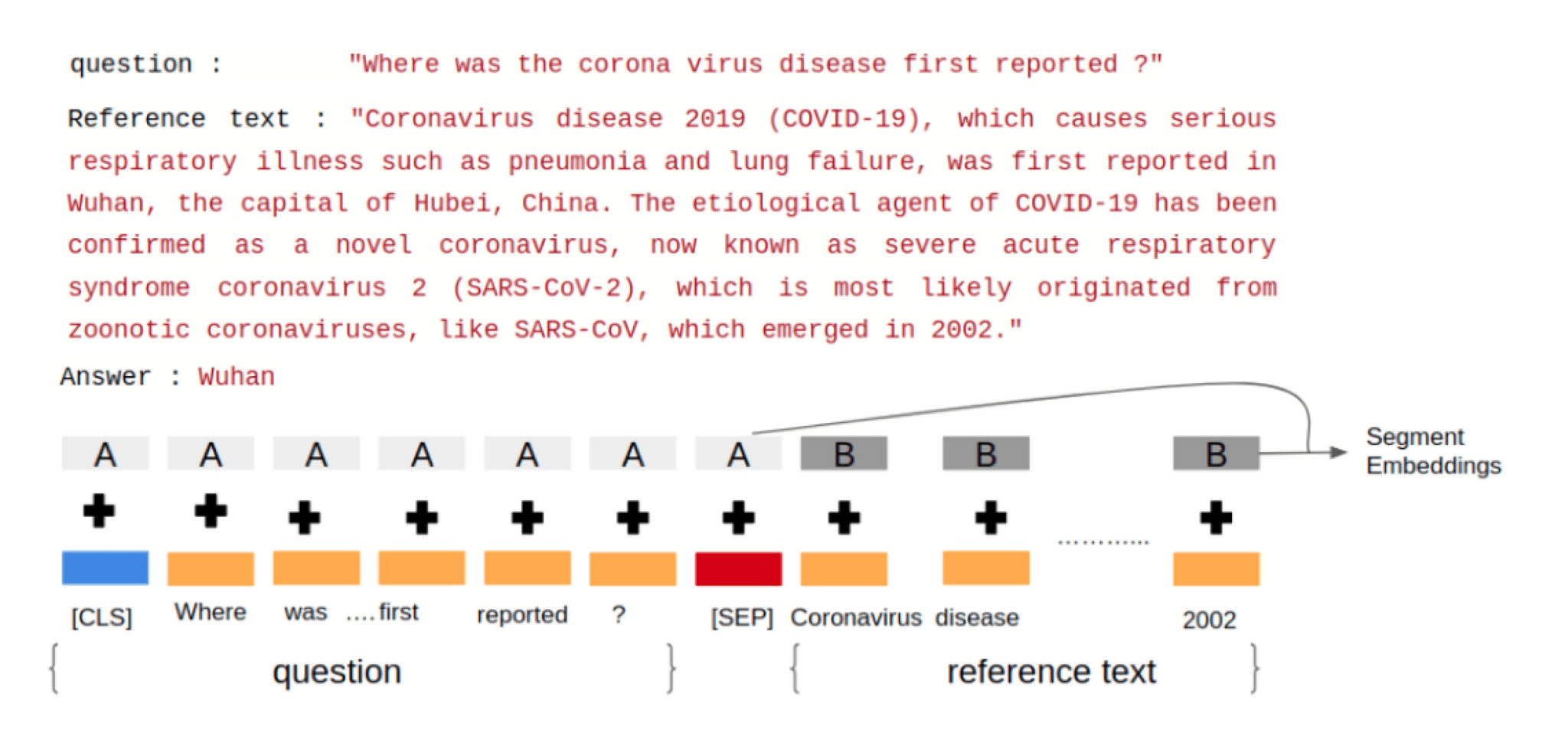
To feed a QA task into BioBERT, we pack both the question and the reference text into the input tokens. The two pieces of text are separated by the special [SEP] token. BioBERT also uses “Segment Embeddings” to differentiate the question from the reference text. We also add a classification [CLS] token at the beginning of the input sequence. A positional embedding is also added to each token to indicate its position in the sequence. We then tokenized the input using word piece tokenization technique [3] using the pre-trained tokenizer vocabulary. Any word that does not occur in the vocabulary (OOV) is broken down into sub-words greedily. For example, if play, ##ing, and ##ed are present in the vocabulary but playing and played are OOV words then they will be broken down into play + ##ing and play + ##ed respectively (## is used to represent sub-words). The input is then passed through 12 transformer layers at the end of which the model will have 768-dimensional output embeddings.
Let us take a look at an example to understand how the input to the BioBERT model appears. Consider the research paper “Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19)“ [6] from Pubmed. We use the abstract as the reference text and ask the model a question to see how it tries to predict the answer to this question. Figure 2 explains how we input the reference text and the question into BioBERT.
Start & End Token Classifiers
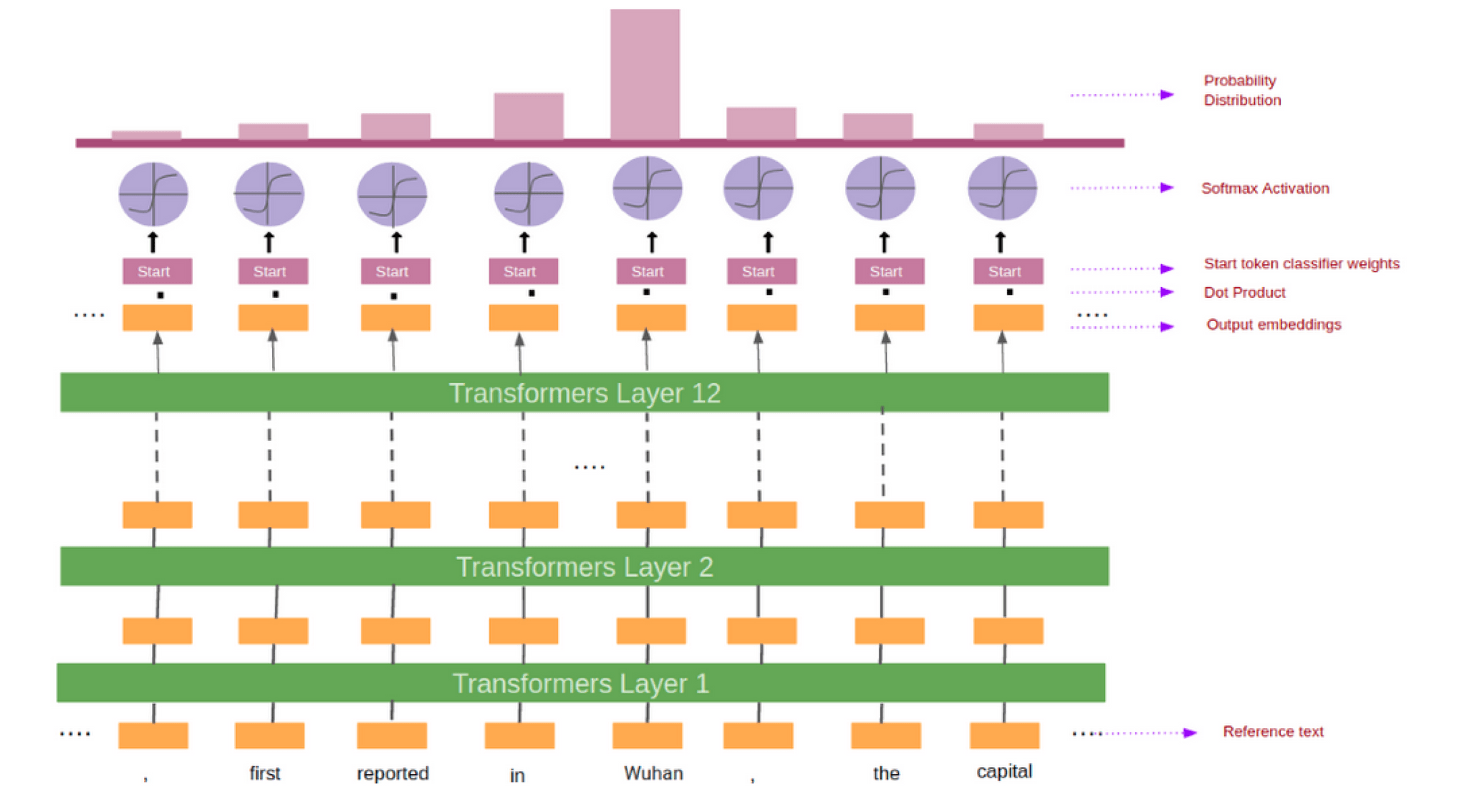
BioBERT needs to predict a span of a text containing the answer. This is done by predicting the tokens which mark the start and the end of the answer. For every token in the reference text we feed its output embedding into the start token classifier.
After taking the dot product between the output embeddings and the start weights (learned during training), we applied the softmax activation function to produce a probability distribution over all of the words. Whichever word has the highest probability of being the start token is the one that we picked. We repeat this process for the end token classifier. Figure 3 shows the pictorial representation of the process.
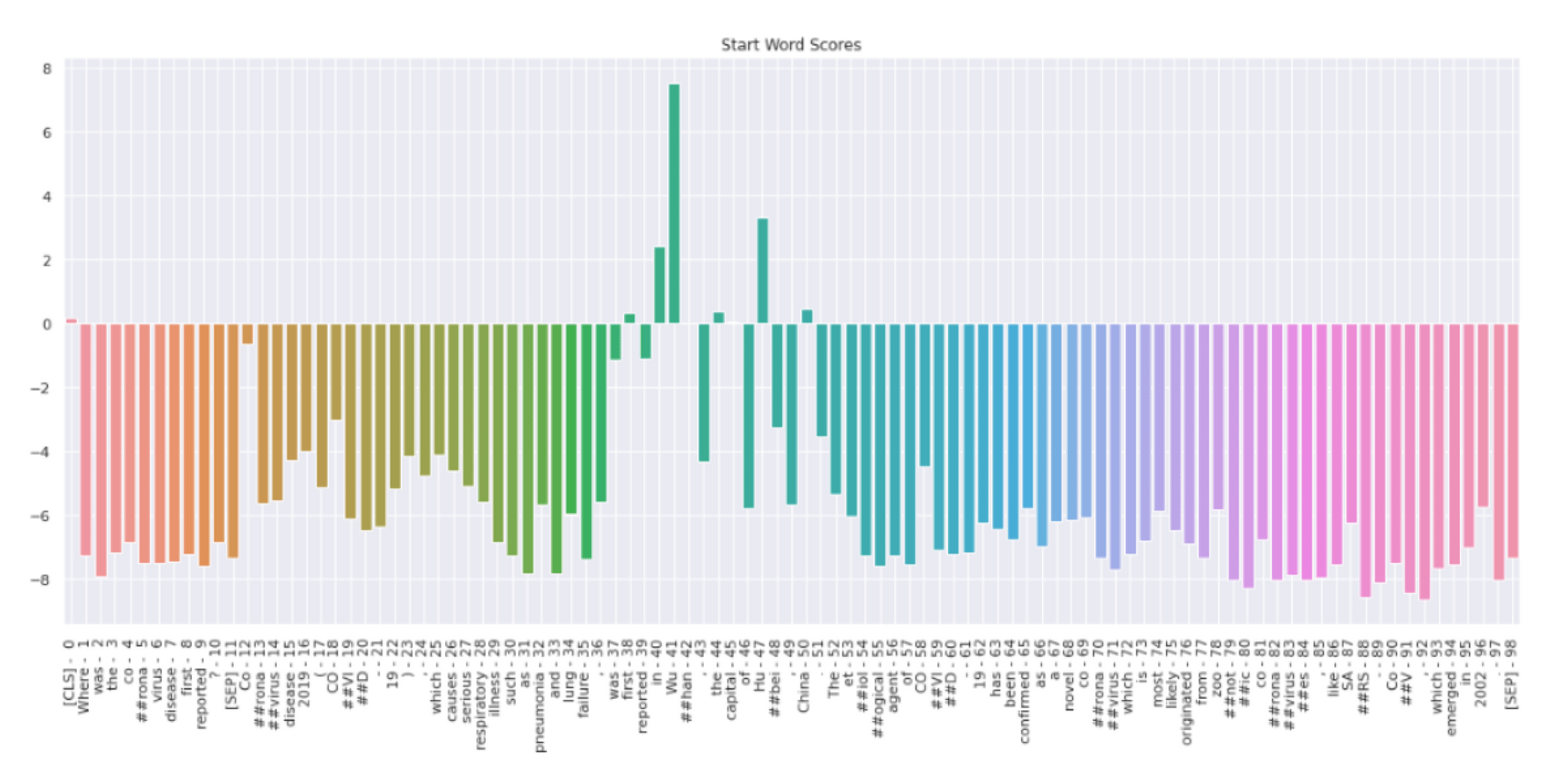
In figure 4, we can see the probability distribution of the start token. Token “Wu” has the highest probability score followed by “Hu”, and “China”. All the other tokens have negative scores. Therefore, the model predicts that Wu is the start of the answer.
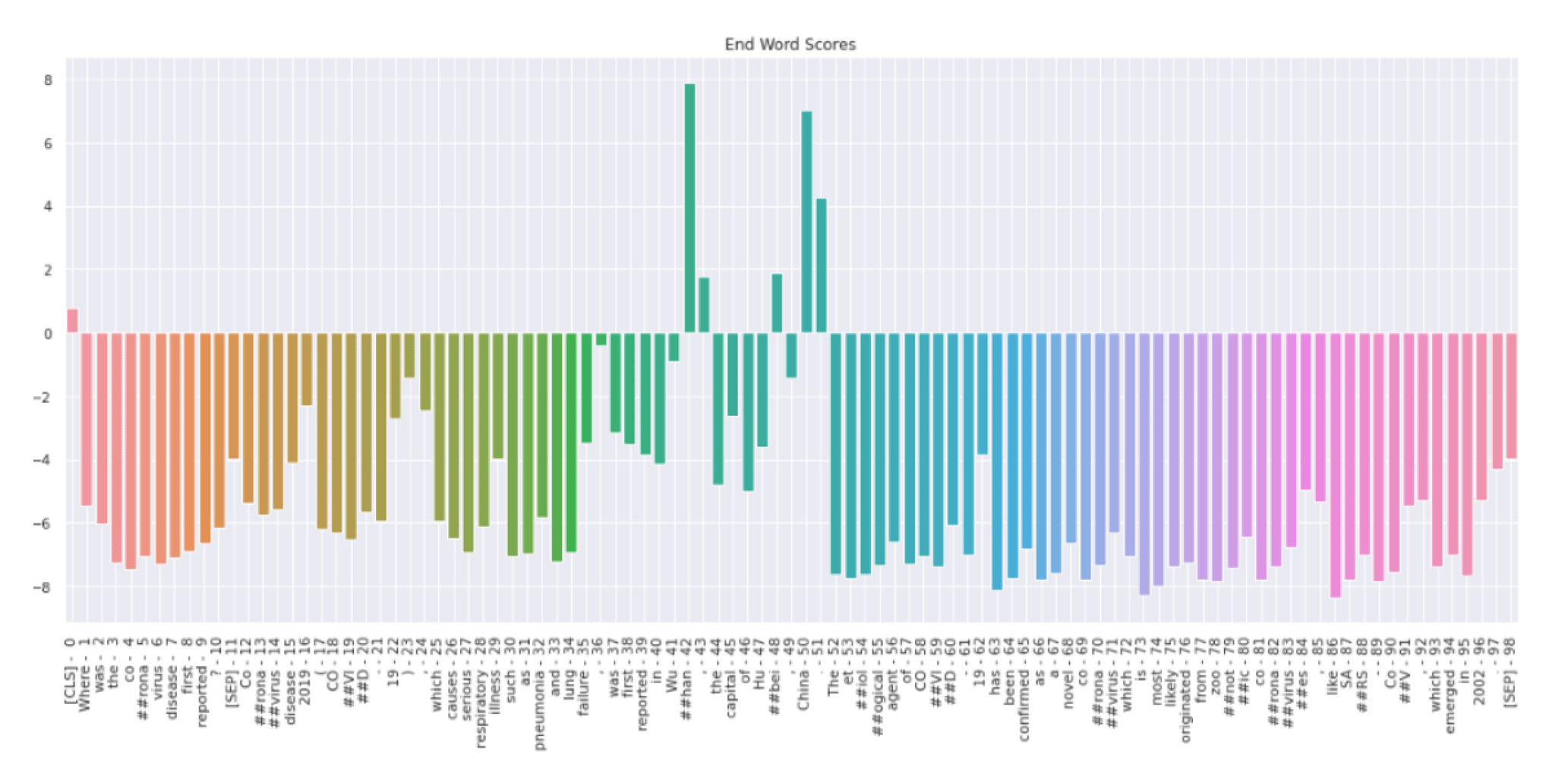
In figure 5, we can see the probability distribution of the end token. Token “##han” has the highest probability score followed by “##bei” and “China”. All other tokens have negative scores. Therefore, the model predicts that ##han is the end of the answer. Model thus predicts Wuhan as the answer to the user's question.
Summary and conclusion
Automatic QA systems are a very popular and efficient method for automatically finding answers to user questions. We have presented a method to create an automatic QA system using doc2vec and BioBERT that answers user factoid questions. I hope this article will help you in creating your own QA system.
References
[2] Le Q, Mikolov T. Distributed representations of sentences and documents. InInternational conference on machine learning 2014 Jan 27 (pp. 1188-1196).
[3] Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020 Feb 15;36(4):1234-40.
[4] Rajpurkar P, Jia R, Liang P. Know what you don't know: Unanswerable questions for SQuAD. arXiv preprint arXiv:1806.03822. 2018 Jun 11.
[5] Staff CC. Cs 224n default final project: Question answering on squad 2.0. Last updated on February. 2019;28.
[6] Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, Kim BT, Kim SJ. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19).